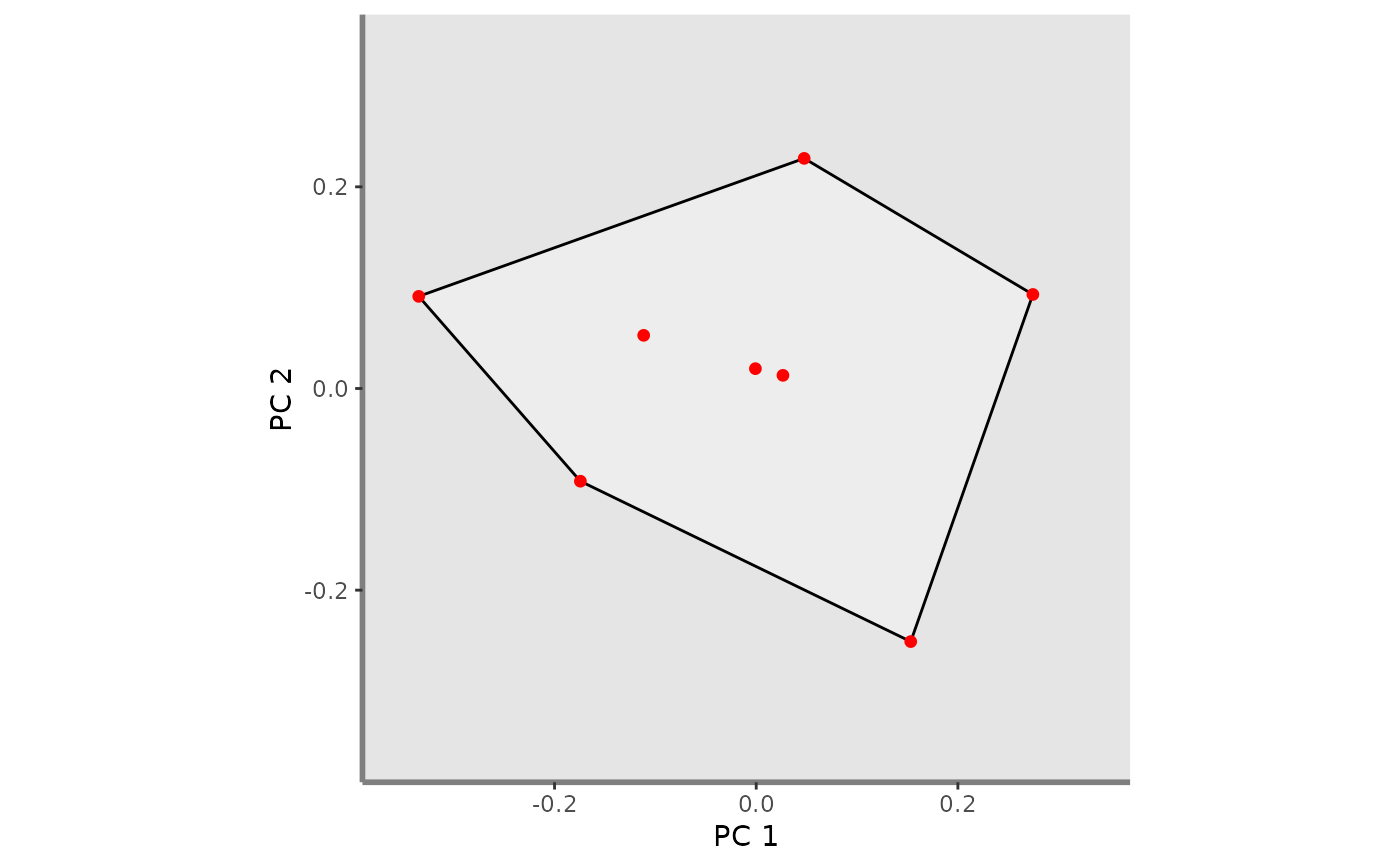
This function plots FRic index for a given pair of functional axes and one or several assemblages. It adds convex hull(s), points and vertices of 1:N assemblages on the background plot
Usage
fric.plot(
ggplot_bg,
asb_sp_coord2D,
asb_vertices_nD,
plot_sp = TRUE,
color_ch,
fill_ch,
alpha_ch,
shape_sp,
size_sp,
color_sp,
fill_sp,
shape_vert,
size_vert,
color_vert,
fill_vert
)Arguments
- ggplot_bg
a ggplot object of the plot background retrieved through the
background.plotfunction.- asb_sp_coord2D
a list of matrix (ncol = 2) with coordinates of species present in each assemblage for a given pair of axes for a given pair of functional axes.
- asb_vertices_nD
a list (with names as in asb_sp_coord2D) of vectors with names of species being vertices in n dimensions.
- plot_sp
a logical value indicating whether species of each assemblage should be plotted or not. Default: plot_sp = TRUE.
- color_ch
a R color name or an hexadecimal code referring to the color of the border of the convex hull filled by the pool of species if one assemblage to plot or a vector of R color names or hexadecimal codes if several assemblages to plot. If more than one assemblage to plot, the vector should be formatted as: c(asb1 = "firstRcolorname", asb2 = "secondRcolorname", ...).
- fill_ch
a R color name or an hexadecimal code referring to the color of convex hull filling if one assemblage to plot or a vector of R color names or hexadecimal codes if several assemblages to plot. If more than one assemblage to plot, the vector should be formatted as: c(asb1 = "firstRcolorname", asb2 = "secondRcolorname", ...).
- alpha_ch
a numeric value referring to the value of transparency of the convex hull filling (0 = high transparency, 1 = no transparency) if one assemblage to plot or a vector numeric values if several assemblages to plot. If more than one assemblage to plot, the vector should be formatted as: c(asb1 = "firstRtransparency", asb2 = "secondRtransparency", ...).
- shape_sp
a numeric value referring to the shape of the symbol used for species plotting if one assemblage to plot or a vector numeric values if several assemblages to plot. If more than one assemblage to plot, the vector should be formatted as: c(asb1 = "firstRshape", asb2 = "secondRshape", ...).
- size_sp
a numeric value referring to the size of the symbol used for species plotting if one assemblage to plot or a vector numeric values if several assemblages to plot. If more than one assemblage to plot, the vector should be formatted as: c(asb1 = "firstRsize", asb2 = "secondRsize", ...).
- color_sp
a R color name or an hexadecimal code referring to the color of species if one assemblage to plot or a vector of R color names or hexadecimal codes if several assemblages to plot. If more than one assemblage to plot, the vector should be formatted as: c(asb1 = "firstRcolorname", asb2 = "secondRcolorname", ...).
- fill_sp
a R color name or an hexadecimal code referring to the color of species symbol filling (if
shape_sp> 20) if one assemblage to plot or a vector of R color names or hexadecimal codes if several assemblages to plot. If more than one assemblage to plot, the vector should be formatted as: c(asb1 = "firstRcolorname", asb2 = "secondRcolorname", ...).- shape_vert
a numeric value referring to the shape of the symbol used for vertices plotting if one assemblage to plot or a vector numeric values if several assemblages to plot. If more than one assemblage to plot, the vector should be formatted as: c(asb1 = "firstRshape", asb2 = "secondRshape", ...).
- size_vert
a numeric value referring to the size of the symbol used for vertices plotting if one assemblage to plot or a vector numeric values if several assemblages to plot. If more than one assemblage to plot, the vector should be formatted as: c(asb1 = "firstRsize", asb2 = "secondRsize", ...).
- color_vert
a R color name or an hexadecimal code referring to the color of vertices if one assemblage to plot or a vector of R color names or hexadecimal codes if several assemblages to plot. If more than one assemblage to plot, the vector should be formatted as: c(asb1 = "firstRcolorname", asb2 = "secondRcolorname", ...). If color_vert = NA, vertices are not plotted (for shapes only defined by color, ie shape inferior to 20. Otherwise
fillmust also be set to NA).- fill_vert
a R color name or an hexadecimal code referring to the color of vertices symbol filling (if
shape_vert>20) if one assemblage to plot or a vector of R color names or hexadecimal codes if several assemblages to plot. If more than one assemblage to plot, the vector should be formatted as: c(asb1 = "firstRcolorname", asb2 = "secondRcolorname", ...). Iffill = NAandcolor = NA, vertices are not plotted (ifshape_vertsuperior to 20
Examples
# Load Species*Traits dataframe:
data("fruits_traits", package = "mFD")
# Load Assemblages*Species dataframe:
data("baskets_fruits_weights", package = "mFD")
# Load Traits categories dataframe:
data("fruits_traits_cat", package = "mFD")
# Compute functional distance
sp_dist_fruits <- mFD::funct.dist(sp_tr = fruits_traits,
tr_cat = fruits_traits_cat,
metric = "gower",
scale_euclid = "scale_center",
ordinal_var = "classic",
weight_type = "equal",
stop_if_NA = TRUE)
#> [1] "Running w.type=equal on groups=c(Size)"
#> [1] "Running w.type=equal on groups=c(Plant)"
#> [1] "Running w.type=equal on groups=c(Climate)"
#> [1] "Running w.type=equal on groups=c(Seed)"
#> [1] "Running w.type=equal on groups=c(Sugar)"
#> [1] "Running w.type=equal on groups=c(Use,Use,Use)"
# Compute functional spaces quality to retrieve species coordinates matrix:
fspaces_quality_fruits <- mFD::quality.fspaces(sp_dist = sp_dist_fruits,
maxdim_pcoa = 10,
deviation_weighting = "absolute",
fdist_scaling = FALSE,
fdendro = "average")
# Retrieve species coordinates matrix:
sp_faxes_coord_fruits <-
fspaces_quality_fruits$details_fspaces$sp_pc_coord
# Retrieve species coordinates matrix for the assemblage "basket_1":
sp_filter <- mFD::sp.filter(asb_nm = c("basket_1"),
sp_faxes_coord = sp_faxes_coord_fruits,
asb_sp_w = baskets_fruits_weights)
sp_faxes_coord_fruits_b1 <- sp_filter$`species coordinates`
# Reduce it to the two studid axes: PC1 and PC2:
sp_faxes_coord_fruits_b1_2D <- sp_faxes_coord_fruits_b1[, c("PC1", "PC2")]
# Set faxes limits:
# set range of axes if c(NA, NA):
range_sp_coord_fruits <- range(sp_faxes_coord_fruits)
range_faxes_lim <- range_sp_coord_fruits + c(-1, 1)*(range_sp_coord_fruits[2] -
range_sp_coord_fruits[1]) * 0.05
# Retrieve the background plot:
ggplot_bg_fruits <- mFD::background.plot(
range_faxes = range_faxes_lim,
faxes_nm = c("PC 1", "PC 2"),
color_bg = "grey90")
# Retrieve vertices names:
vert_nm_fruits <- vertices(sp_faxes_coord_fruits_b1_2D,
order_2D = TRUE, check_input = TRUE)
# Plot in white the convex hull of all fruits species:
ggplot_fric <- mFD::fric.plot(
ggplot_bg = ggplot_bg_fruits,
asb_sp_coord2D = list(basket_1 = sp_faxes_coord_fruits_b1_2D),
asb_vertices_nD = list(basket_1 = vert_nm_fruits),
plot_sp = TRUE,
color_ch = c("basket_1" = "black"),
fill_ch = c("basket_1" = "white"),
alpha_ch = c("basket_1" = 0.3),
size_sp = c("basket_1" = 1),
shape_sp = c("basket_1" = 16),
color_sp = c("basket_1" = "red"),
fill_sp = c("basket_1" = "red"),
size_vert = c("basket_1" = 1),
color_vert = c("basket_1" = "red"),
fill_vert = c("basket_1" = "red"),
shape_vert = c("basket_1" = 16))
ggplot_fric